Epigenetic Mechanisms: Emerging Therapeutic Targets for Blood Disorders
Despite the emergence of highly effective therapies for certain blood cancers, others remain incurable today. There is a critical need to identify new approaches and treatments that do not rely on existing paradigms and strategies. Toward this goal, the sequencing of the human genome has not only revolutionized fundamental scientific knowledge, but also catalyzed efforts to elucidate the genetic basis of human disease and provided a powerful platform to forge a new paradigm of precision medicine.
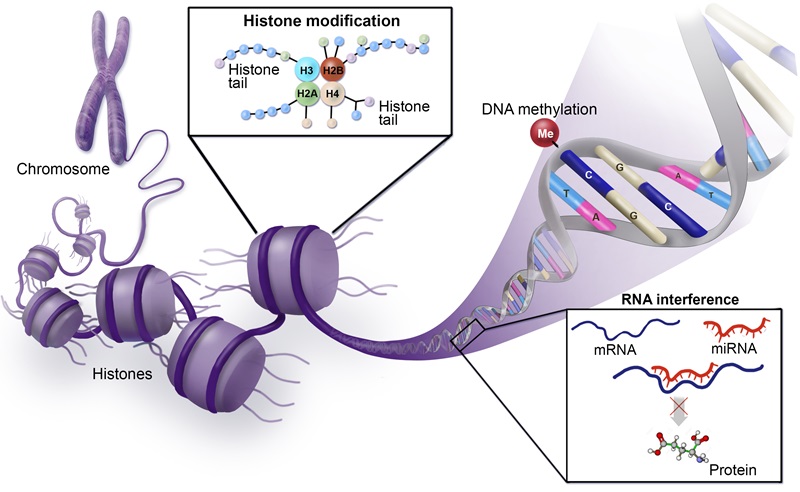
While the impact of genomics on science and human health cannot be overestimated, DNA sequence variation represents only one part of the complex regulation of gene function. Epigenetics, which encompasses alterations in gene expression caused by mechanisms other than DNA sequence changes, also plays a significant role in phenotypic diversity and differential sensitivity to environmental stimuli. However, unraveling these epigenetic mechanisms represents an extraordinarily challenging problem, given the vast number of molecular constituents involved. Chromatin, which is produced as DNA wraps around histone octamers to form nucleosomes, plays a significant role in genome regulation by dictating whether genes are active in a given cell or tissue type and is subject to extensive post-translational modifications that define this activity.
As the field of epigenetics has advanced, it has become increasingly clear that epigenetic mechanisms provide a completely new ensemble of therapeutic targets for treating hematologic disorders – both non-malignant and malignant. Studies on epigenetic mechanisms have already helped identify a battery of druggable enzymes that represent potential treatment targets that may modulate cell function. In addition, these mechanisms have enormous implications for understanding the molecular underpinnings of the normal orderly development of the nascent hematopoietic system in the embryo as well as hematologic disorders – both non-malignant and malignant, and both constitutional and somatic.
Targets with Transformative Potential: Priorities for Epigenetic Research in Hematologic Disease
Research on epigenetics, including its mechanisms and drivers, will elucidate highly valuable targets and lead to potentially transformative treatment regimens for hematologic disorders.

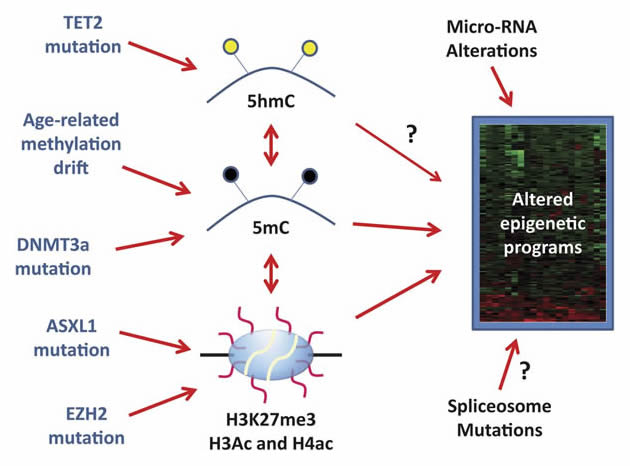
One of the greatest challenges in effectively treating hematologic disorders is the diversity of molecular abnormalities that underlie a disease. However, many common threads are emerging, including alterations in proteins that function through epigenetic mechanisms. For example, in AML with the 3q21;q26 cytogenetic abnormality (which has a particularly poor prognosis), a segment of a chromosome is inverted, placing a DNA sequence that normally activates its neighboring gene in close proximity to a cancer-causing proto-oncogene. This elevates expression of the proto-oncogene, disrupting the epigenetic mechanism controlling cell proliferation and therefore driving leukemogenesis. Similar mechanisms have been noted in other hematologic disorders, including leukemias, lymphomas, multiple myeloma, and MDS.
Since alterations in epigenetic mechanisms commonly mediate hematologic malignancies, epigenetic regulators represent a rich pipeline of potential targets that could be transformative for the treatment of blood cancers. Potential epigenetic targets include proteins that control histone methylation and acetylation, which can regulate genes that control proliferation; proteins that read histone marks and orchestrate changes in gene expression that cause malignancy; and DNA methyltransferases. For instance, the state of the epigenome determines the accessibility of DNA to the transcriptome machinery, thus allowing DNA to execute its functions. Further, fundamental mechanistic studies will continue to lead to the discovery of promising new targets.
Additional research will lay the groundwork in the context of precision medicine, elucidating potentially critical determinants of responsiveness to therapeutic regimens.
| 1.1 | Targeting and reversing malignant histone modifications. Enzymes that catalyze addition and removal of post-translational modifications to histone tails are recurrently mutated in a variety of blood cancers. Many researchers have shown that the abnormal distribution of the histone marks resulting from mutation of these enzymes can activate or silence expression of critical genes involved in malignant transformation. Finding ways to reverse these pathogenic histone marks, either through inhibiting the writers and erasers of the specific mark or by direct epigenetic reprogramming, could have a beneficial effect in restoring normal gene expression programs and alleviating the disease. |
| 1.2 | Identifying small molecules that target epigenetic modifiers in hematopoietic malignancies. Genetic mutations that produce gain-of-function or neomorphic proteins (such as IDH1/2 mutations, EZH2Y641F) are common in leukemias and lymphomas. The mutant proteins go on to corrupt the epigenome resulting in oncogenic gene expression signatures. Development of small molecules that specifically inhibit the mutant protein, but not the wild-type (these mutations are typically heterozygous meaning the wild-type protein is still expressed from the non-mutated allele), could prove beneficial for many patients. |
| 1.3 | Developing tools and technologies for locus-specific epigenetic reprogramming. Current epigenetic therapies (e.g., hypomethylating agents) cause global changes to the epigenome, but the disease is likely driven by specific, local epigenetic dysregulation at several critical loci. The development of tools such as modified CRISPR/Cas9 systems to target and correct aberrant DNA methylation patterns at specific disease-causing genes while leaving the rest of the normal epigenetic marks in the cell intact (for translational purposes), warrants further investigation. |
Studies on epigenetic mechanisms have extraordinary promise to transform therapeutic approaches for non-malignant hematologic disorders. Although the identification of the hemoglobin beta-chain mutation in sickle cell disease represented the first discovery of the genetic basis for disease, there has been limited progress in advancing therapies to counteract the often-crippling complications of this condition. Recently, powerful technologies have been developed with new lines of investigation that aim to transform sickle cell disease and thalassemia therapeutics.
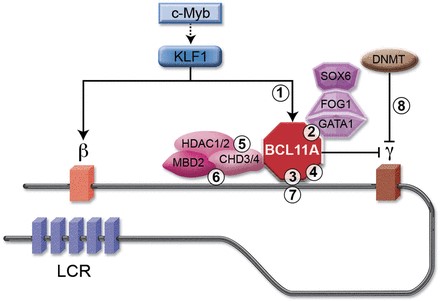
For example, elevating gamma-globin can suppress the toxic effects of mutant beta-globin that underlie sickle cell disease phenotypes. An ensemble of proteins has been implicated in mediating the epigenetic repression of gamma-globin expression, raising the possibility that antagonizing the actions of these proteins to increase gamma-globin expression may be a useful treatment strategy for sickle cell disease. In certain cases, the respective proteins are deemed “undruggable,” based on their structural attributes, so considerably more work is required to identify druggable components of these multi-step epigenetic mechanisms.
There is a critical need to develop better models that will more effectively predict efficacy in humans and better assays that can identify modulators of “undruggable” proteins. Major efforts are required to develop therapeutic approaches for proteins already identified, as well as newly discovered targets. Given the rich proteome and improved technologies available today, studies of proteomics, metabolomics, and regulatory RNAs are likely to reveal promising translational avenues. Answering the many remaining questions on epigenetic mechanisms will require additional research focused on:
| 2.1 | Identifying optimal approaches to use the Bcl11a mechanism for altering globin gene expression. |
| 2.2 | Further understanding of alternative epigenetic pathways and strategies to modulate globin gene expression. |
| 2.3 | Advancing knowledge of the basic mechanisms that regulate the globin genes, which may lead to the development of new classes of agents. For example, investigation of the three-dimensional structure of the nucleus and how it affects gene regulation is a new frontier that may lead to novel therapeutic strategies. |